Tuberculosis (TB) is the leading cause of morbidity and mortality in the HIV and non-HIV immunocompromised populations. TB primarily targets the respiratory system but can disseminate to other organs in the body. Disseminated TB (DTB), also known as miliary TB (MTB), is characterized by the systemic spread of the disease through tiny millet-sized lesions (1–4 mm) that can rapidly develop in various organs. DTB has become increasingly diagnosed in adults due to the growing prevalence of immunocompromising conditions such as diabetes, malignancy, and HIV/AIDS infections, as well as the increased use of immunosuppressive medications (Abuabat et al., 2023; Shafer et al., 1991; Sharma et al., 2012, 2016; Hassanein and Elbadry, 2016).
Diagnosing DTB is challenging due to nonspecific symptoms, atypical presentations, difficulties getting patient samples, and false-negative findings, leading to a delay in the diagnosis and increased mortality (Sharma et al., 2012; Hassanein and Elbadry, 2016). DTB has received less research attention than other forms of TB, at least in part because mycobacterial blood cultures are unavailable in most high-burden settings and have limited diagnostic value because median time to positivity is longer than median time from admission to death in fatal cases of DTB.
Metagenomic next-generation sequencing (mNGS) has been used to identify specific infectious pathogens in a timely, unbiased, and hypothesis-free manner (Zhang et al., 2014; Pang et al., 2024; Wu et al., 2024; Li et al., 2023; Liu et al., 2023; Gao et al., 2024; Shi et al., 2021; Zhou et al., 2019). It also has the ability to recognize co-infecting microbes. Furthermore, research indicates that mNGS can detect hematologic dissemination using blood tests, which may obviate the need for invasive procedures (Pang et al., 2024; Wu et al., 2024; Zhou et al., 2019). In recent years, accumulating data showed that mNGS is effective for diagnosing TB in direct clinical samples with excellent accuracy and specificity (Li et al., 2023; Liu et al., 2023; Gao et al., 2024; Shi et al., 2021; Zhou et al., 2019). Nevertheless, no research assessed the diagnostic accuracy of mNGS in patients with DTB. In this study, we explored the diagnostic efficacy of mNGS in DTB patients using blood samples.
Patients and methodsThis retrospective analysis consecutively enrolled 48 suspected DTB patients admitted to the Second Xiangya Hospital, Central South University (Changsha, China) from 1 May 2021 to 31 May 2024. Patients were eligible for enrollment if they met all the following criteria: (1) consent to undergo the plasma mNGS examination and (2) suspected DTB. Patients were excluded if they met any of the following criteria: (1) incomplete medical record and (2) patients already receiving antituberculosis therapy. Suspected DTB patients were included if they met at least one of the following criteria: (1) miliary pattern on chest imaging and (2) suspected bloodstream infections and TB exposure history. The final clinical diagnostic criteria for DTB include the following: (1) identifying either a miliary pattern on chest imaging and a positive MTB in sputum (culture, acid-fast staining smears, Xpert, or NGS) or lung biopsy showing granulomas, (2) active TB clinical manifestations and positive plasma mNGS results for MTB, or (3) detecting caseating granulomas in a biopsy from any other organ, along with the presence of a miliary pattern on chest imaging or a positive sputum culture for MTB. In the absence of microbiological evidence, the attending physician may diagnose DTB infection clinically by relating the patient’s clinical manifestations and imaging results to exclude other diseases, together with the patient’s confirmed responsiveness to anti-TB treatment after 1 month of follow-up. This study was approved by the Research Ethics Committee of the Second Xiangya Hospital, and the need for written informed consent was waived owing to its retrospective design.
Clinical data collectionData were collected from the patients’ medical records. The following details at diagnosis were gathered: demographics, presenting symptoms, imaging, and histopathological data, as well as risk factors and mortality rates. Mycobacterial blood culture and TB polymerase chain reaction (PCR)-based assays of blood samples cannot be collected due to the limitations of routine methods. In cases of incomplete data, historical manual files were retrieved and examined.
Detection method (mNGS)Specimens were obtained from patients according to standard procedures. Briefly, 3 mL of blood was collected in sterile DNase-free tubes. Plasma was separated by centrifuging the blood at 1,600 × g at 4°C for 10 min within 8 h of collection. Using high-throughput sequencing technology, the DNA sequence of the microorganisms in the sample was obtained and compared with that of the microbial gene bank to identify pathogenic microorganisms. The detection process includes nucleic acid extraction, library construction, computer sequencing, bioinformatic analysis, and report interpretation (Liu et al., 2023). The detection process was based on the Illumina NextSeq550Dx (Illumina) sequencing system, and reference databases for classification were downloaded from NCBI. RefSeq contains 11,958 bacterial genomes or scaffolds, 1,714 fungi related to human infection, 7,373 whole-genome sequences of viral taxa, and 343 parasites associated with human diseases. The number of TB reads was defined as the number of unique reads for the standardized species. MTB was considered positive when the mapping read number (genus or species level) was in the top 20 in the bacteria list (Miao et al., 2018).
Statistical analysisContinuous data are deemed as nonparametric. Continuous variables were expressed as median and interquartile range (IQR), and the Shapiro–Wilk test (p < 0.05) was used for continuous variables that exhibited non-normal distribution. The categorized data were represented by case number (n) and percentage (%). The chi-square test (McNemar test) was employed for comparison. p < 0.05 was statistically significant without adjustment for multiple tests. All analyses were conducted with the IBM SPSS statistical software, version 26.0.
ResultsPatient profilesAmong 48 individuals with suspected DTB infection, 20 were identified as non-DTB cases (Figure 1). A total of 28 cases were confirmed DTB. Table 1 summarizes the demographic characteristics of the 28 enrolled individuals diagnosed with DTB. The median age was 55 years, nearly 50% were over 60 years old, and 17 individuals (60.7%) were male. The predominant risk factor was HIV infection, present in 10 (35.7%) patients. Diabetes or pneumoconiosis was observed in four patients (14.3%); chronic liver disease was detected in 7.14% of patients; chronic kidney disease, anorexia, or pregnancy was observed in 3.6% of patients; and four (14.3%) patients had immunocompromising conditions, including malignancy or use of immunosuppressive agents. The most common presenting symptoms observed were fever, fatigue, and weight loss, occurring in 24 (85.7%), 20 (71.4%), and 23 (82.1%) patients, respectively. Cough was reported in 10 (35.7%) patients, night sweats in 14 (50%) patients, and hepatosplenomegaly in 16 (57.1%) patients. Regarding the anatomical site of TB infection at the time of diagnosis, the majority of patients had pulmonary MTB, which accounted for over half of the cases (16 patients; 57.1%). TB lymphadenitis and gastrointestinal involvement were the second most common sites, observed in six patients (21.4%), followed by pleural or peritoneal involvement in four patients(14.3%); and urogenital TB in one patient (3.6%).
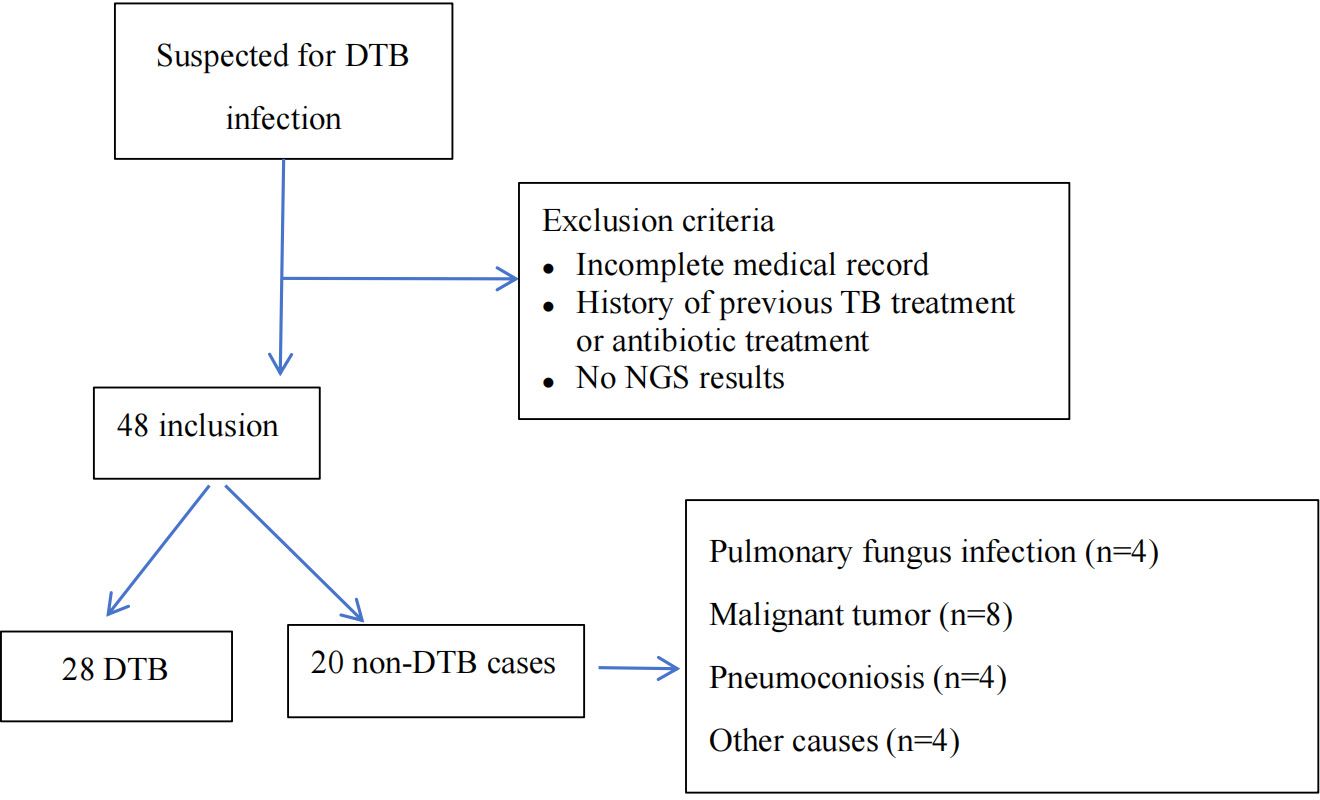
Figure 1. Flowchart of enrollment, exclusion criteria, and study procedures, by mNGS blood assay result. DTB, disseminated TB; NGS, next-generation sequencing.
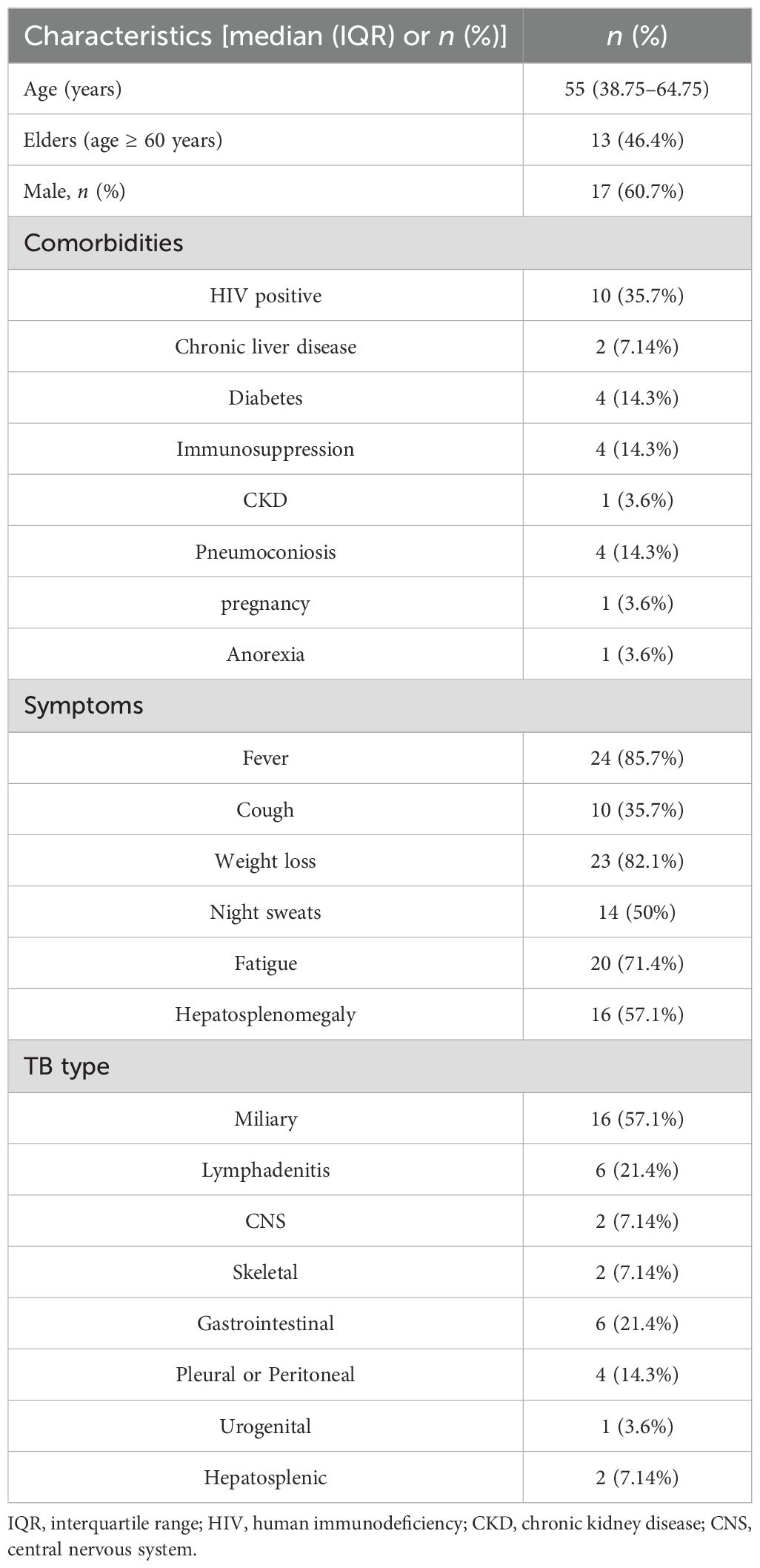
Table 1. Demographic characteristics.
Table 2 outlines the most frequently observed laboratory findings in the study population. The most common laboratory abnormality was elevated ESR, present in 26 (92.9%) patients. Anemia was the second most prevalent laboratory finding, observed in 25 (89.3%) patients. Elevated liver enzymes were also frequently observed, with aspartate aminotransferase elevated in 12 (42.9%) patients, and alanine transaminase elevated in 11 (39.3%) patients. Leukopenia was present in 2 (7.14%) patients, thrombocytopenia in 4 (14.3%) patients, thrombocytosis or leukocytosis in 7 (25%) patients, and elevated creatinine in 7 (25%) patients. Positive T-SPOT.TB results were reported in 23 (82.1%) patients. CD4 T-cell subsets in the peripheral blood were analyzed in 22 individuals, of whom 16 (72.7%) showed a reduction. mNGS of blood samples was conducted on all patients; 19 (67.9%) contained TB sequences, with specific reads of TB ranging from 1 to 219. TB sequences were identified in 15 patients (53.6%) with over two reads and in 12 patients (42.9%) with over three reads. A total of 18 acid-fast bacilli (AFB) smears from sputum, pleural, or ascitic fluid were performed, of which 8 showed positive results. Bronchoalveolar lavage was performed five times, with two positive results. X-pert MTB/RIF was taken 14 times, with 6 positive outcomes. Positive biopsy results were observed in four patients with caseating granuloma.
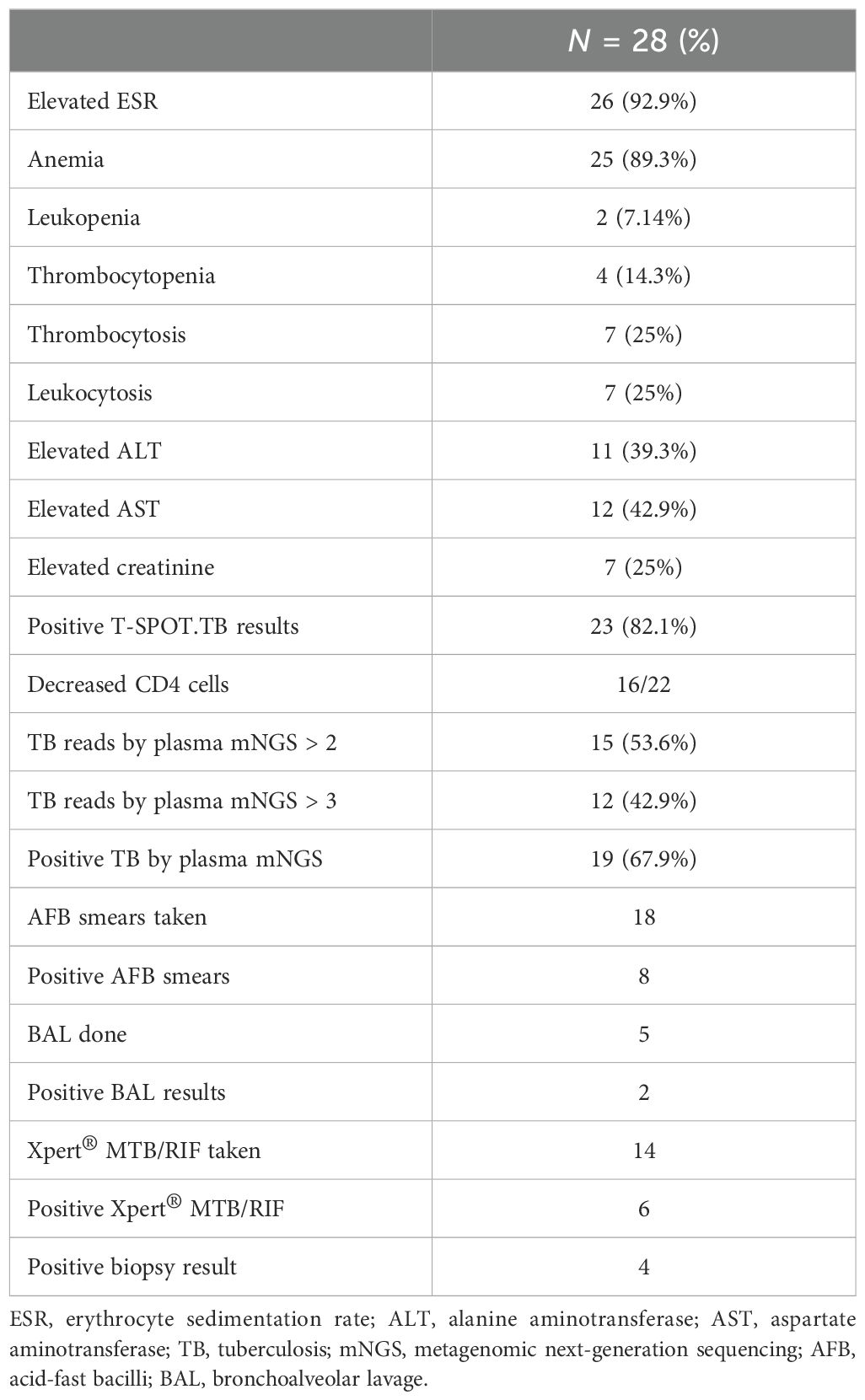
Table 2. Laboratory findings.
Comparison of clinical features of DTB in patients with HIV infection and non-HIV infectionTable 3 assesses the clinical characteristics of 18 DTB persons with non-HIV infections and 10 DTB patients with HIV infections. The HIV-positive group exhibits elevated TB mNGS readings (p = 0.012) and TB mNGS sensitivity (p = 0.05) compared to the non-HIV group. The sensitivity of TB mNGS in blood samples was 80% for HIV-infected patients and 44.4% for non-HIV-infected individuals (p = 0.05). The non-HIV cohort had a higher prevalence of MTB (p = 0.018), but extrapulmonary TB was more prevalent in the HIV-positive group. An older age was observed, although it lacked statistical significance in the non-HIV groups. No significant differences were seen between the two groups in regard to AST, ALT, WBC, HB, PLT, CRP, PCT, and Cr levels.
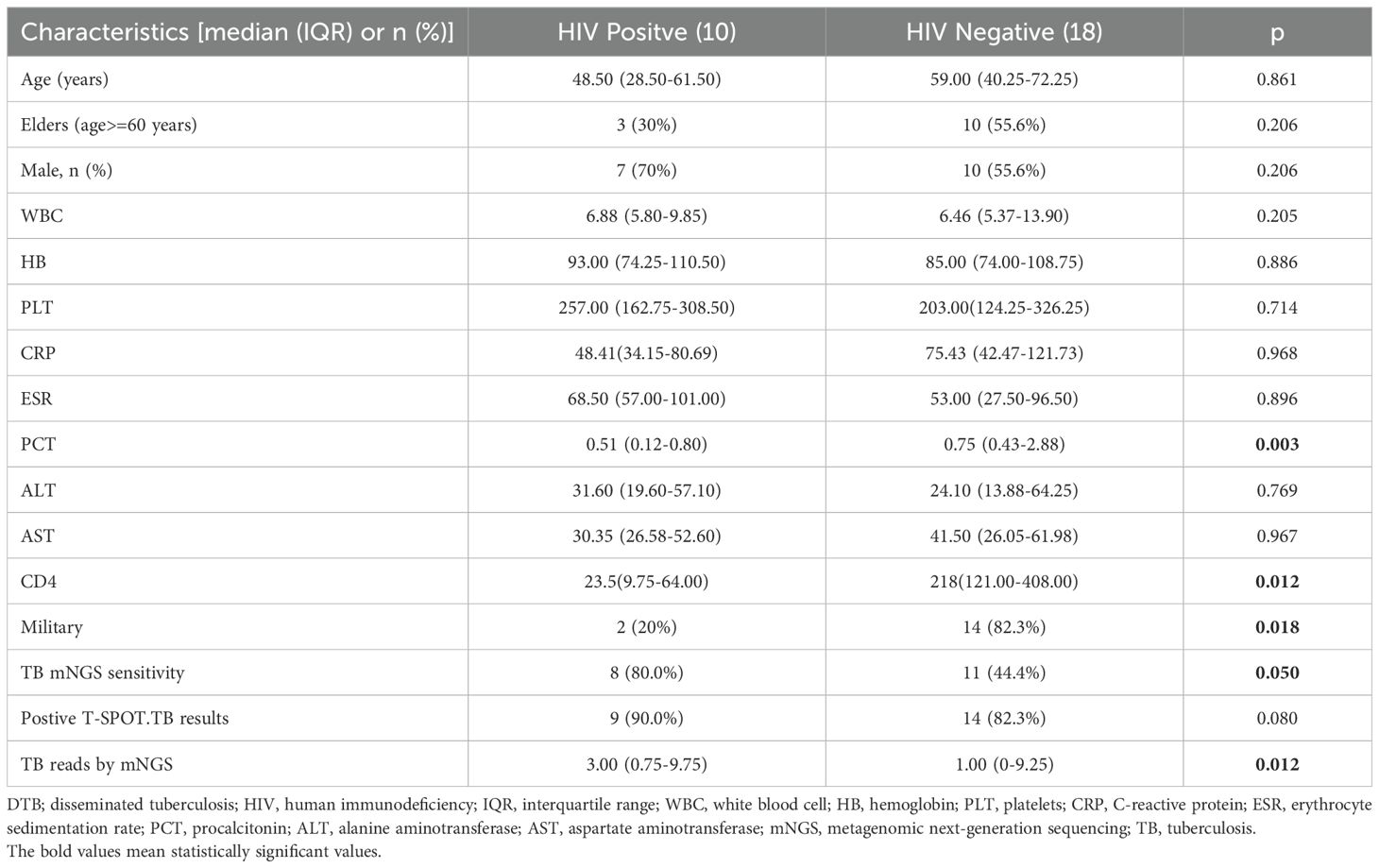
Table 3. Comparison of clinical features of DTB in patients with HIV infection and non-HIV infection.
Comparison of clinical features of DTB in mNGS-positive and -negative patients with TBTable 4 illustrates that mNGS of blood samples was performed in all patients; the diagnostic sensitivity of mNGS was calculated using the clinical composite diagnosis as reference standard. TB was identified by mNGS in 19 (67.9%) cases. The sensitivity of mNGS was 67.9%. In a comparison of TB mNGS-positive and -negative groups, TB sequences were more detectable by mNGS in male patients, those with elevated PCT levels, those who are HIV positive, and those with a decreased CD4 T-cell count. There were no significant differences between the two groups in terms of age, AST, ALT, WBC, HB, PLT, CRP, and Cr levels.
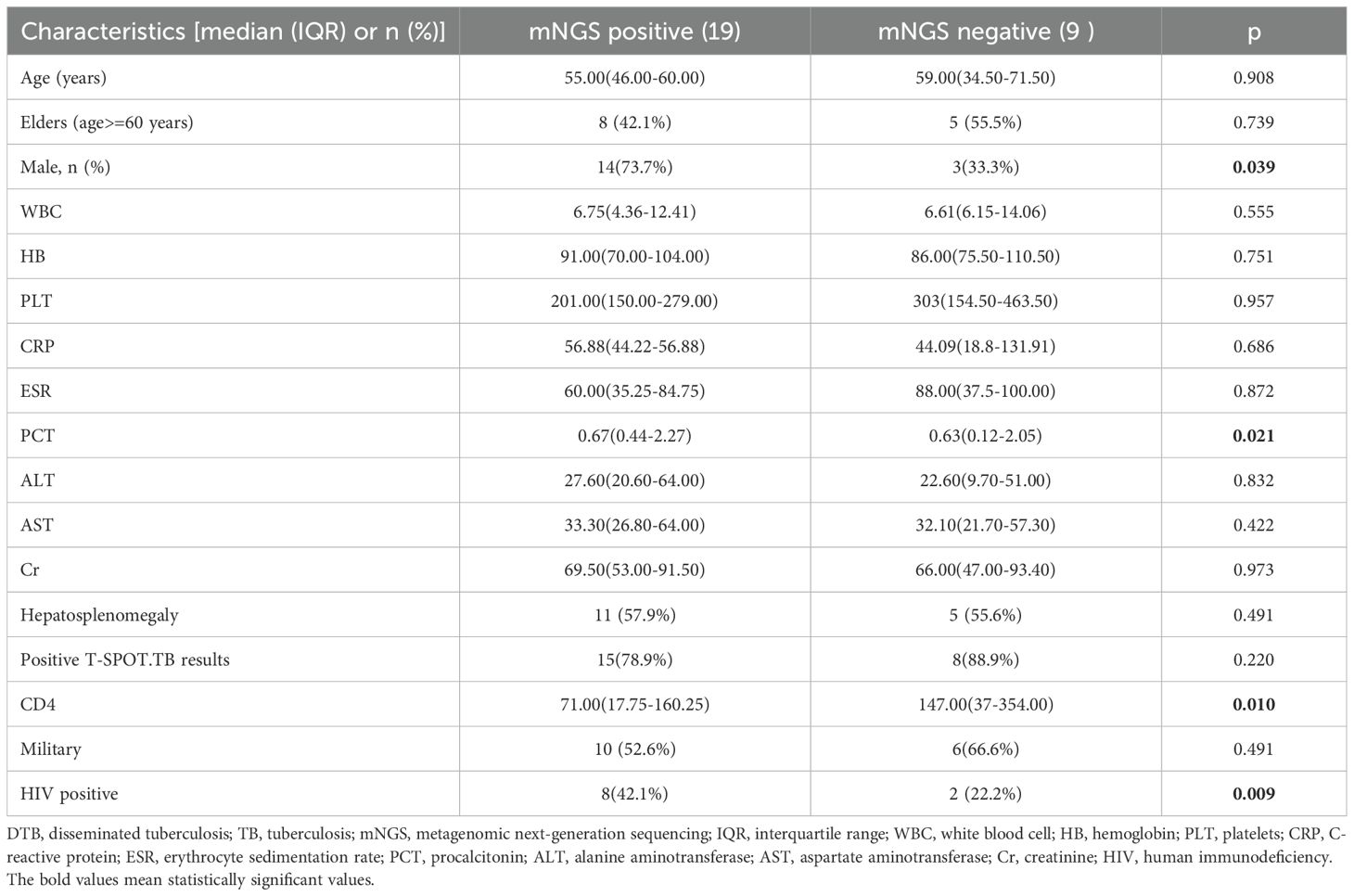
Table 4. Comparison of clinical features of DTB in mNGS-positive and mNGS-negative patients with TB.
Diagnostic performance of blood mNGS and T-SPOT.TB assay in confirmed DTBWe assessed the diagnostic performance for blood mNGS and T-SPOT.TB methods compared with the clinical final diagnosis of DTB (Figure 2). Of the 28 patients with confirmed DTB, 23 cases were detected by T-SPOT.TB, with a positive rate of 82.1%, while mNGS found 19 cases, with a positive rate of 67.8%. Among the 19 patients with positive mNGS, 4 patients had negative T-SOPT.TB assays, whereas the remaining 15 patients had both positive mNGS and T-SPOT.TB assay. In these 28 patients, the positive rates of mNGS and TSPOT were not (p > 0.05) statistically different. With clinically confirmed DTB as the reference standard, mNGS demonstrated a clinical sensitivity of 67.8%, a clinical specificity of 100%, a positive predictive value of 100%, and a negative predictive of 69.0%. With clinically verified DTB as the reference standard, the T-SPOT.TB assay had a clinical sensitivity of 82.1%, a clinical specificity of 80%, a positive predictive value of 85.2%, and a negative predictive value of 76.2%.
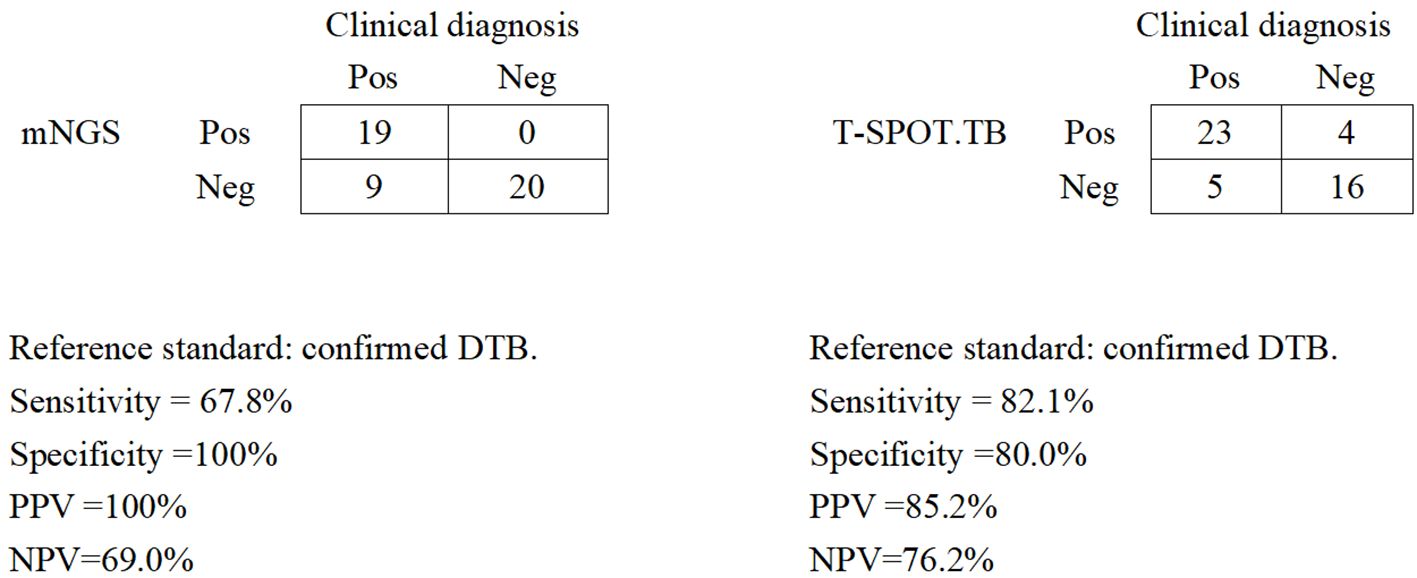
Figure 2. Diagnostic performance of mNGS and the T-SPOT.TB assay for DTB using blood samples. Pos, positive; Neg, negative; mNGS, metagenomic next-generation sequencing; DTB, disseminated tuberculosis; PPV, positive predictive value; NPV, negative predictive value.
DiscussionIn light of the continuing HIV/AIDS epidemic and increasing use of immunosuppressive and cytotoxic drugs, the burden of DTB will continue to rise (Meira et al., 2019). DTB is a notoriously difficult disease to diagnose, with nonspecific symptoms and clinical abnormalities that are shared by many other infectious and noninfectious diseases. Mortality from this disease has remained high despite effective therapy being available. The early, timely, and correct diagnosis of DTB is a key factor for reducing mortality. This study applied mNGS of blood samples to evaluate its diagnostic efficacy for DTB and chose the use of clinical diagnosis rather than mycobacterial blood culture as a comparison because of the limitation of routine methods. Despite the lack of culture for clinical diagnosis, pathological findings and follow-up greatly reduced the possibility of misdiagnosis.
This study revealed HIV infection as the most prevalent risk factor for DTB, accounting for 35.7%; other risk factors were pharmacological immunosuppression, diabetics, chronic liver disease, and chronic kidney disease. Consistent with those reported in the literature (Ye et al., 2021; Crump et al., 2012; Kerkhoff et al., 2017), the importance of cell-mediated immunity was further illustrated by the inverse correlation found between the CD4+ T-cell count in HIV patients and the frequency of cases of DTB. Previous studies have highlighted the prognostic value of T-cell count in this context (Ye et al., 2021; Crump et al., 2012).
Several investigations from Africa with blood culture have shown that TB is a common cause of bloodstream infections (Crump et al., 2012; Kerkhoff et al., 2017). Blood is an attractive sample type for diagnosing DTB, especially for HIV-infected individuals, due to their higher probability of sputum culture-negative, disseminated, or extrapulmonary TB. Various studies on Mycobacterium tuberculosis bacteremia have established the potential of blood for TB detection by both culture and nucleic acid amplification test (NAAT) (Lima et al., 2009; Taci et al., 2003).
Mycobacterial blood culture is not available in most settings. Even where available, an average 3-week delay between culture and identification, combined with high early mortality, means that TB blood culture has limited diagnostic value. NAAT-based M. tuberculosis bacteremia studies have had different results, with sensitivities of TB detection in peripheral blood of 2% to 55% (Lima et al., 2009; Taci et al., 2003; Rebollo et al., 2006). Patients with DTB are at high risk for early death, and this sequential approach to empirical therapy introduces a potentially fatal delay in the commencement of anti-TB chemotherapy (ATC). Rapid testing of blood for TB with the mNGS may facilitate the rapid diagnosis of DTB and consequently the early initiation of ATC. Our findings revealed that 19 blood samples contained TB sequences from 28 diagnoses as DTB, indicating that plasma mNGS had a high sensitivity (67.9%) and that circulating cfDNA sequencing contributed to an improvement in the diagnosis of DTB. Our results demonstrated that TB sequence was more detectable by plasma mNGS in patients with HIV infection, elevated PCT levels, and decreased CD4 T-cell count. Patients who were HIV positive had higher TB mNGS reads and TB mNGS sensitivity. The sensitivity of TB mNGS in blood samples was 80% for HIV-infected patients and 44.4% for non-HIV-infected individuals. The data also revealed that immunocompromised patients were more likely to yield positive mNGS results, and detection of M. tuberculosis bacteremia by mNGS was rapid and had high specificity (100%) and high sensitivity (67.9%) compared to blood culture and NAAT.
In the diagnostic specificity of DTB, mNGS is better than T-SPOT.TB, although the T-SPOT.TB assay is slightly more sensitive than mNGS for DTB; a cost/time trade-off would be beneficial. In China, the mNGS test costs between $300 and $450 per sample, whereas the cost of the TSOPT.TB assay ranges from approximately $84 to $100. The detection period for mNGS is 1 day, while the detection time for TSPOT.TB assay takes approximately 48 h, and research indicates that mNGS can identify drug-resistant genes or mixed pathogens, altering the therapeutic management of TB patients and reducing the cost-effectiveness (Brown et al.,2015; Doyle et al.,2018; Votintseva et al., 2017).
mNGS is not recommended for routine use in mild/moderate infections due to its high cost (Chiu and Miller, 2019; Miller et al., 2018). Nonetheless, it is necessary to carry out adequate development in complex patients. MTB is an intracellular bacterium, and its DNA extraction is challenging, which may result in reduced susceptibility to mNGS. Moreover, the false positives of mNGS should be carefully monitored and assessed. MTB is not a common background bacteria in laboratory settings; the possibility for contamination of MTB in blood samples is lower than that in other sample types. Therefore, even when one specific read of a taxon is mapped to either the species or genus level, MTB infection should be considered. MTB was considered positive if it meets the following criteria: (1) its genus or species level is among the top 20 with the highest standardized specific reading number (SDSMRN); (2) it ranks first within its genus; and (3) its SDSMRN exceeds 1. To reduce the risk of contamination introduced before mNGS procedures, we made a strict sterile procedure during sample collection and a relative nucleic acid-free guideline during specimen preparation. In addition, two independent experienced clinics analyzed the mNGS results to see whether they were in accordance with the patient’s clinical manifestation and diagnosis.
There are some limitations to this study. First, the sample size included was relatively small, comprising only 28 cases. Thus, the sensitivity of diagnosis DTB for mNGS may not be accurately evaluated, M. tuberculosis bloodstream infection is underdiagnosed, false negatives will increase, and the negative predictive value could be overestimated. Prospective studies with a bigger sample size are needed to further evaluate the diagnostic value of mNGS in DTB. Second, the patients included did not undergo traditional culture; comparative analysis with conventional methods was limited by lack of information; therefore, prospective studies are needed to compare mNGS with traditional detection methods. Third, biases resulting from the selection and referral processes are potentially important, since these investigations were conducted in single centers. Despite some limitations, this investigation provides useful insights into the clinical value of plasma mNGS for identifying DTB. A comprehensive, multicenter study is needed to verify our findings.
In conclusion, our study showed that mNGS of blood samples exhibits exceptional sensitivity for diagnosing DTB. The TB sequence was more detectable by peripheral blood mNGS in patients who were HIV positive or with other immunosuppressive diseases. Blood mNGS should be conducted when diagnosing DTB is challenging, particularly in areas with a high prevalence of HIV presenting with severe sepsis in order to prevent the high early death rate. However, further refinement and larger-scale validation studies are necessary, especially in HIV-positive or other immunosuppressive populations.
Data availability statementThe raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statementThe studies involving humans were approved by Ethics Committee of the Second Xiangya Hospital of the Central South University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributionsJM: Writing – original draft. YJ: Formal analysis, Funding acquisition, Writing – review & editing. YH: Formal analysis, Data curation, Supervision, Writing – review & editing. HZ: Conceptualization, Project administration, Writing – review & editing.
FundingThe author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Scientific Research Program of FuRong Laboratory (No. 2023SK2108), the Clinical Medical Research Center for Viral Hepatitis in Hunan Province (No. 2023SK4009), and the Hunan Provincial Health Commission Research Program (No. 202303088786).
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ReferencesAbuabat, F., Badri, M., Abuabat, S. (2023). Sultan Alsultan, Salim Baharoon, Abdullah Alharbi,et al; Disseminated tuberculosis: Clinical presentation, diagnosis, and outcomes in a tertiary-care hospital in Saudi Arabia. Int. J. Mycobacteriol 12, 407–411. doi: 10.4103/ijmy.ijmy_141_23
PubMed Abstract | Crossref Full Text | Google Scholar
Brown, A. C., Bryant, J. M., Einer-Jensen, K. (2015). Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples, et al; Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples. J. Clin. Microbiol. 53, 2230–2237. doi: 10.1128/JCM.00486-15
PubMed Abstract | Crossref Full Text | Google Scholar
Crump, J. A., Ramadhani, H. O., Morrissey, A. B., Morrissey, A. B., Saganda, W., Mwako, M. S., et al. (2012). Bacteremic disseminated tuberculosis in sub-saharan Africa: a prospective cohort study. Clin. Infect. Dis. 55, 242—250. doi: 10.1093/cid/cis409
PubMed Abstract | Crossref Full Text | Google Scholar
Doyle, R. M., Burgess, C., Williams, R., Gorton, R., Booth, H., Brown, J., et al. (2018). Direct whole-genome sequencing of sputum accurately identifies drugresistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J. Clin. Microbiol. 56, e00666–e00618. doi: 10.1128/JCM.00666-18
PubMed Abstract | Crossref Full Text | Google Scholar
Gao, J., Zhao, L., Chen, G., Huang, C., Kong, W., Feng, Y., et al. (2024). The value of metagenomic next-generation sequencing for the diagnosis of pulmonary tuberculosis using bronchoalveolar lavage fluid. Lab. Med. 55, 96–102. doi: 10.1093/labmed/lmad041
PubMed Abstract | Crossref Full Text | Google Scholar
Hassanein, H. A., Elbadry, M. I. (2016). Selective immunoglobulin M deficiency in an adult with miliary tuberculosis: A clinically interesting coexistence. A case report and review of the literature. Int. J. Mycobacteriol. 5, 106–110. doi: 10.1016/j.ijmyco.2015.11.002
PubMed Abstract | Crossref Full Text | Google Scholar
Kerkhoff, A. D., Barr, D. A., Schutz, C., Burton, R., Nicol, M. P., Lawn, S. D., et al. (2017). Disseminated tuberculosis among hospitalised HIV patients in South Africa: a common condition that can be rapidly diagnosed using urine-based assays. Sci. Rep. 7, 10931. doi: 10.1038/s41598-017-09895-7
PubMed Abstract | Crossref Full Text | Google Scholar
Li, Y., Bian, W., Wu, S., Zhang, J., Li, D. (2023). Metagenomic next-generation sequencing for Mycobacterium tuberculosis complex detection: a meta-analysis. Front. Public. Health 11. doi: 10.3389/fpubh.2023.1224993
PubMed Abstract | Crossref Full Text | Google Scholar
Lima, J., Montenegro, L., de Albuquerque, M., Cabral, M., Lima, A., Abath, F., et al. (2009). Performance of nested PCR in the specific detection of Mycobacterium tuberculosis complex in blood samples of pediatric patients. J. Bras. Pneumol. 35, 690–6697. doi: 10.1590/S1806-37132009000700011
PubMed Abstract | Crossref Full Text | Google Scholar
Liu, Y., Wang, H., Li, Y., Yu, Z. (2023). Clinical application of metagenomic next-generation sequencing in tuberculosis diagnosis. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.984753
PubMed Abstract | Crossref Full Text | Google Scholar
Meira, L., Chaves, C., Araújo, D., Almeida, L., Boaventura, R., Ramos, A., et al. (2019). Predictors and outcomes of disseminated tuberculosis in an intermediate burden setting. Pulmonology 25, 320–327. doi: 10.1016/j.pulmoe.2018.11.001
PubMed Abstract | Crossref Full Text | Google Scholar
Miao, Q., Ma, Y., Wang, Q., Pan, J., Zhang, Y., Jin, W., et al. (2018). Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin. Infect. Dis. 67, S231–S240. doi: 10.1093/cid/ciy693
PubMed Abstract | Crossref Full Text | Google Scholar
Miller, J. M., Binnicker, M. J., Campbell, S., Campbell, S., Carroll, K. C., Chapin, K. C., et al. (2018). A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the infectious diseases society of America and the American society for microbiology. Clin. Infect. Dis. 67, e1–e94. doi: 10.1093/cid/ciy381
PubMed Abstract | Crossref Full Text | Google Scholar
Pang, F., Xu, W., Zhao, H., Chen, S., Tian, Y., Fu, J., et al. (2014). Comprehensive evaluation of plasma microbial cell-free DNA sequencing for predicting bloodstream and local infections in clinical practice: a multicenter retrospective study.Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1256099
PubMed Abstract | Crossref Full Text | Google Scholar
Rebollo, M. J., San Juan Garrido, R., Folgueira, D., Palenque, E., DiazPedroche, C., Lumbreras, C., et al. (20062006). Blood and urine samples as useful sources for the direct detection of tuberculosis by polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 56, 141–146. doi: 10.1016/j.diagmicrobio.2006.03.018
PubMed Abstract | Crossref Full Text | Google Scholar
Shafer, R. W., Kim, D. S., Weiss, J. P., Quale, J. M. (1991). Extrapulmonary tuberculosis in patients with human immunodeficiency virus infection. Med. (Baltimore) 70, 384–397. doi: 10.1097/00005792-199111000-00004
PubMed Abstract | Crossref Full Text | Google Scholar
Sharma, S. K., Mohan, A., Sharma, A. (2012). Challenges in the diagnosis & treatment of miliary tuberculosis. Indian. J. Med. Res. 135, 703–730.
Shi, Y. F., Shi, X. H., Zhang, Y., Chen, J. X., Lai, W. X., Luo, J. M., et al. (2021). Disseminated tuberculosis associated hemophagocytic lymphohistiocytosis in a pregnant woman with Evans syndrome: A case report and literature review. Front. Immunol. 12. doi: 10.3389/fimmu.2021.676132
PubMed Abstract | Crossref Full Text | Google Scholar
Taci, N., Yurdakul, A. S., Ceyhan, I., Berktas, M. B., Ogretensoy, M. (2003). Detection of Mycobacterium tuberculosis DNA from peripheral blood in patients with HIV-seronegative and new cases of smear-positive pulmonary tuberculosis by polymerase chain reaction. Respir. Med. 97, 676–6681. doi: 10.1053/rmed.2003.1500
PubMed Abstract | Crossref Full Text | Google Scholar
Votintseva, A. A., Bradley, P., Pankhurst, L., Elias, C. D. O., Loose, M., Nilgiriwala, K., et al. (2017). Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J. Clin. Microbiol. 55, 1285–1298. doi: 10.1128/JCM.02483-16
PubMed Abstract | Crossref Full Text | Google Scholar
Wu, J., Song, W., Yan, H., Luo, C., Hu, W., Xie, L., et al. (2024). Metagenomic next-generation sequencing in detecting pathogens in pediatric oncology patients with suspected bloodstream infections. Pediatr. Res. 95, 843–851. doi: 10.1038/s41390-023-02776-y
PubMed Abstract | Crossref Full Text | Google Scholar
Ye, Y., Yang, N., Zhou, J., Qian, G., Chu, J. (2021). Case report: metagenomic next-generation sequencing in diagnosis of disseminated tuberculosis of an immunocompetent patient. Front. Med. (Lausanne) 8. doi: 10.3389/fmed.2021.687984
PubMed Abstract | Crossref Full Text | Google Scholar
Zhang, H., Zhou, F., Liu, X., Huang, J. (2014). Clinical application of metagenomic next-generation sequencing in patients with different organ system infection: A retrospective observational study. Med. (Baltimore). 103, e36745. doi: 10.1097/MD.0000000000036745
PubMed Abstract | Crossref Full Text | Google Scholar
Zhou, X., Wu, H., Ruan, Q., Luo, C., Hu, W., Xie, l., et al. (2019). Clinical Evaluation of Diagnosis Efficacy of Active Mycobacterium tuberculosis Complex Infection via Metagenomic Next-Generation Sequencing of Direct Clinical Samples. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00351
留言 (0)